 Technology peripherals
Technology peripherals
 AI
AI
 Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis
Over the past two years, machine learning has revolutionized protein structure prediction. Now, artificial intelligence has triggered a new round of revolution in the field of protein design.
Since the advent of AI, many scientists have joined the track of using it to conduct protein research. Biologists have discovered that using machine learning, protein molecules can be created in seconds. In the past, this time might have been several months.
Recently, the start-up Evozyne used pre-trained AI models provided by NVIDIA to create two proteins with significant potential in the medical and clean energy fields. One of the proteins is used to treat a congenital disease, and another is used to consume carbon dioxide to reduce global warming.
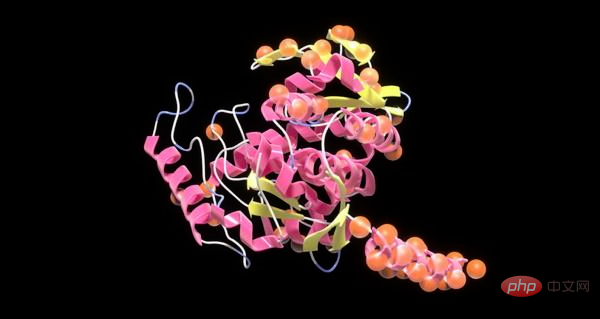
So scientists use NVIDIA BioNeMo to create large language models that generate high-quality proteins to speed up drug development and help create a more sustainable environment.
New ways to accelerate drug discovery
Andrew Ferguson, co-founder of Evozyne and co-author of the paper, said: “It is gratifying that this AI model has produced the first round of production. The resulting synthetic proteins are just like naturally occurring proteins, indicating that the model has learned nature's design rules."
Evozyne uses NVIDIA's ProtT5. ProtT5 is a Transformer model that is part of NVIDIA BioNeMo, a software framework and service for creating medical AI models.
Molecular engineer Ferguson, whose research fields include chemistry and machine learning, said: "BioNeMo is very powerful and allows us to train a model and then use the model to run work tasks at a very low cost, in a matter of seconds. Millions of sequences can be generated in minutes. The model predicts how to assemble new proteins that meet Evozyne's needs."
The model is at the heart of the Evovyne ProT-VAE pipeline. Evozyne’s ProT-VAE pipeline combines the powerful Transformer model in NVIDIA BioNeMo with variational autoencoders (VAEs).
He said: "A few years ago, no one noticed that you could use large language models combined with variational autoencoders to design proteins."
In contrast, Evozyne's method can change half or more of the amino acids in a protein in just one round. This amounts to hundreds of mutations.
Evozyne data scientist Joshua Moller said: "They speed up training by extending the work to multiple GPUs.
This reduces the time to train large AI models from months to a week "So we can train models that would otherwise be impossible to train," Ferguson said, "such as models with billions of trainable parameters. ”
Revolutionary AI Model
The traditional protein engineering design method, namely directed evolution, uses a slow, unplanned approach, usually just one protein at a time. Change the sequence of a few amino acids. Machine learning helps to study a large number of possible amino acid combinations and then effectively identify the most useful sequences.
BioNeMo is an AI-enabled drug development built on NVIDIA NeMo Megatron Cloud service and framework for training and deploying large biomolecule Transformer AI models at supercomputing scale. Services include pre-trained LLM, native support for common file formats for proteins, DNA, RNA and chemistry, and also provides support for SMILES ( Data loader used by (for molecular structures) and FASTA (for amino acid and nucleotide sequences).
With BioNeMo, scientists can start easily using pre-trained models, automatic downloaders and Preprocessor. With unsupervised structured learners, various models, embeddings, and outputs are combined to combine multimodal data. Unsupervised pretraining also eliminates the need for labeled data, allowing for rapid generation Learned embeddings predict protein structure, function, cellular location, water solubility, membrane binding, conservation and variable regions, etc.
Among them, MegaMolBART is a method that uses 1.4 billion molecules (SMILES string ), which can be used for a variety of chemical informatics applications. Moreover, BioNeMo provides Transformer-based protein language models such as ProtT5 and ESM1-85M.
BioNeMo also provides OpenFold, which is a user-friendly A deep learning model for predicting the 3D structure of novel protein sequences.
NVIDIA's Transformer model reads the amino acid sequences in millions of proteins. The model uses the technology used by neural networks to understand text and learns how to How nature builds protein amino acid sequences.
Looking to the future, the prospects of using AI to accelerate protein engineering are very broad. Artificially designed proteins are more stable than proteins that originally exist in nature and can survive without energy or One of its functions can also be realized under extreme conditions such as high temperature.
In addition, artificial intelligence can also be used to design amino acid sequences to match the backbone, which can be used to improve the stability of specific proteins such as enzymes and antibodies. Artificial intelligence technology plays a very important role in the design of proteins of different sizes and different conformations. In the future, it can also help design more and more useful proteins, including new biological materials that can be used to reduce pollution and improve the environment.
The above is the detailed content of Endless Possibilities NVIDIA Generative AI Model Speeds Protein Synthesis. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1664
1664
 14
14
 1422
1422
 52
52
 1316
1316
 25
25
 1268
1268
 29
29
 1240
1240
 24
24
 How to understand DMA operations in C?
Apr 28, 2025 pm 10:09 PM
How to understand DMA operations in C?
Apr 28, 2025 pm 10:09 PM
DMA in C refers to DirectMemoryAccess, a direct memory access technology, allowing hardware devices to directly transmit data to memory without CPU intervention. 1) DMA operation is highly dependent on hardware devices and drivers, and the implementation method varies from system to system. 2) Direct access to memory may bring security risks, and the correctness and security of the code must be ensured. 3) DMA can improve performance, but improper use may lead to degradation of system performance. Through practice and learning, we can master the skills of using DMA and maximize its effectiveness in scenarios such as high-speed data transmission and real-time signal processing.
 How to use the chrono library in C?
Apr 28, 2025 pm 10:18 PM
How to use the chrono library in C?
Apr 28, 2025 pm 10:18 PM
Using the chrono library in C can allow you to control time and time intervals more accurately. Let's explore the charm of this library. C's chrono library is part of the standard library, which provides a modern way to deal with time and time intervals. For programmers who have suffered from time.h and ctime, chrono is undoubtedly a boon. It not only improves the readability and maintainability of the code, but also provides higher accuracy and flexibility. Let's start with the basics. The chrono library mainly includes the following key components: std::chrono::system_clock: represents the system clock, used to obtain the current time. std::chron
 Quantitative Exchange Ranking 2025 Top 10 Recommendations for Digital Currency Quantitative Trading APPs
Apr 30, 2025 pm 07:24 PM
Quantitative Exchange Ranking 2025 Top 10 Recommendations for Digital Currency Quantitative Trading APPs
Apr 30, 2025 pm 07:24 PM
The built-in quantization tools on the exchange include: 1. Binance: Provides Binance Futures quantitative module, low handling fees, and supports AI-assisted transactions. 2. OKX (Ouyi): Supports multi-account management and intelligent order routing, and provides institutional-level risk control. The independent quantitative strategy platforms include: 3. 3Commas: drag-and-drop strategy generator, suitable for multi-platform hedging arbitrage. 4. Quadency: Professional-level algorithm strategy library, supporting customized risk thresholds. 5. Pionex: Built-in 16 preset strategy, low transaction fee. Vertical domain tools include: 6. Cryptohopper: cloud-based quantitative platform, supporting 150 technical indicators. 7. Bitsgap:
 How to handle high DPI display in C?
Apr 28, 2025 pm 09:57 PM
How to handle high DPI display in C?
Apr 28, 2025 pm 09:57 PM
Handling high DPI display in C can be achieved through the following steps: 1) Understand DPI and scaling, use the operating system API to obtain DPI information and adjust the graphics output; 2) Handle cross-platform compatibility, use cross-platform graphics libraries such as SDL or Qt; 3) Perform performance optimization, improve performance through cache, hardware acceleration, and dynamic adjustment of the details level; 4) Solve common problems, such as blurred text and interface elements are too small, and solve by correctly applying DPI scaling.
 What is real-time operating system programming in C?
Apr 28, 2025 pm 10:15 PM
What is real-time operating system programming in C?
Apr 28, 2025 pm 10:15 PM
C performs well in real-time operating system (RTOS) programming, providing efficient execution efficiency and precise time management. 1) C Meet the needs of RTOS through direct operation of hardware resources and efficient memory management. 2) Using object-oriented features, C can design a flexible task scheduling system. 3) C supports efficient interrupt processing, but dynamic memory allocation and exception processing must be avoided to ensure real-time. 4) Template programming and inline functions help in performance optimization. 5) In practical applications, C can be used to implement an efficient logging system.
 How to measure thread performance in C?
Apr 28, 2025 pm 10:21 PM
How to measure thread performance in C?
Apr 28, 2025 pm 10:21 PM
Measuring thread performance in C can use the timing tools, performance analysis tools, and custom timers in the standard library. 1. Use the library to measure execution time. 2. Use gprof for performance analysis. The steps include adding the -pg option during compilation, running the program to generate a gmon.out file, and generating a performance report. 3. Use Valgrind's Callgrind module to perform more detailed analysis. The steps include running the program to generate the callgrind.out file and viewing the results using kcachegrind. 4. Custom timers can flexibly measure the execution time of a specific code segment. These methods help to fully understand thread performance and optimize code.
 An efficient way to batch insert data in MySQL
Apr 29, 2025 pm 04:18 PM
An efficient way to batch insert data in MySQL
Apr 29, 2025 pm 04:18 PM
Efficient methods for batch inserting data in MySQL include: 1. Using INSERTINTO...VALUES syntax, 2. Using LOADDATAINFILE command, 3. Using transaction processing, 4. Adjust batch size, 5. Disable indexing, 6. Using INSERTIGNORE or INSERT...ONDUPLICATEKEYUPDATE, these methods can significantly improve database operation efficiency.
 How to use string streams in C?
Apr 28, 2025 pm 09:12 PM
How to use string streams in C?
Apr 28, 2025 pm 09:12 PM
The main steps and precautions for using string streams in C are as follows: 1. Create an output string stream and convert data, such as converting integers into strings. 2. Apply to serialization of complex data structures, such as converting vector into strings. 3. Pay attention to performance issues and avoid frequent use of string streams when processing large amounts of data. You can consider using the append method of std::string. 4. Pay attention to memory management and avoid frequent creation and destruction of string stream objects. You can reuse or use std::stringstream.



