 Technology peripherals
Technology peripherals
 AI
AI
 AlphaFold 3 takes an important step towards decoding molecular behavior and biological computing, a Nature sub-journal commented
AlphaFold 3 takes an important step towards decoding molecular behavior and biological computing, a Nature sub-journal commented
AlphaFold 3 takes an important step towards decoding molecular behavior and biological computing, a Nature sub-journal commented

If we fully understood how molecules interact with each other, there would be nothing left to learn about biology, because every biological phenomenon, including how we perceive the world, ultimately originates in cells Behavior and interactions of biomolecules within.
The recently launched AlphaFold 3 can predict the 3D structure of biomolecular complexes directly from the sequences of proteins, nucleic acids and their ligands. This marks significant progress in our long-term exploration of how biomolecules interact.
AlphaFold 3 represents a breakthrough in predicting the three-dimensional structure of a complex directly from its sequence, providing insights into biomolecular interactions.

A one-dimensional (1D) sequence of a biomolecule (such as a protein or nucleic acid) that specifies a cellular function, similar to a piece of code that specifies a program. This sequence represents code in a programming language and is "compiled" into code in machine language through a folding process, forming a unique 3D structure.
- Execution of the program
The program is performed by the interaction between the folded biomolecules and other molecules within the cell.
- Interactions of biomolecules
Due to their unique three-dimensional structure, biomolecules will only interact with a small number of molecules within the cell (such as DNA sites), and these interactions will trigger a series of carefully planned chemical and structural transformations that together define biochemical programs (e.g., transcription). The products of biochemical processes, such as RNA, represent the output of the executing program.
- Encoding of Biological Sequences
Thus, in biology, a one-dimensional sequence of biomolecules encodes the program and the means to compile and execute the program; this sequence encodes the software and hardware. Predicting the three-dimensional structures formed by complexes of biomolecules based on their one-dimensional sequences is a critical step in understanding how biological programs are performed, with profound implications for our ability to understand, rationally manipulate, and design biological systems.

1. AlphaFold 2
- Released in 2020, a revolutionary protein structure prediction algorithm
- Excellent median accuracy Compared with other methods
- provides predicted structures of 200 million known proteins
2. RoseTTAFold
- released in 2021, a protein prediction tool based on deep learning
- prediction accuracy is comparable to AlphaFold 2, faster, Lower computational requirements
- Utilize multi-track neural networks for high accuracy
3. AlphaFold Multimer
- Modified version of AlphaFold 2
- Trained for protein complex datasets
- Improved protein-protein complexes Prediction
4. AlphaFold 3
- Released in May 2023
- Go beyond professional tools to predict the 3D structure of protein complexes
- Significantly improve the prediction accuracy of protein-ligand and protein-nucleic acid complexes
- Predict structures containing multiple covalent modifications
5. Technology update
- Replace the structure module with the diffusion module
- Directly predict the Cartesian coordinates of individual atoms
- Expand to a wider chemical space
Illustration: Illustrative example of the diffusion process that powers AlphaFold 3’s diffusion module. (Source: paper)
As a simplified illustration of AlphaFold 3:
- Imagine taking the three-dimensional coordinates of each atom in a typical biomolecule complex.
- Iteratively add more and more Gaussian noise to it until we get a randomly distributed cloud of atoms in space (forward diffusion).
- Diffusion models use multi-layer neural networks to learn to reverse this process (reverse diffusion).
In this way, the diffusion module in AlphaFold 3 learns to:
- Predict the coordinates of every atom in a given complex without the need for a predefined residue framework.
- Broader chemical space including nucleic acids, ions, ligands and chemical modifications.
Other improvements:
- Replaced Evoformer with Pairformer, a newer Transformer architecture.
- Less emphasis on MSA handling.
- Update metrics to adapt to changes in network architecture.
Progress and Limitations:
- 進步:提高了預測精度,減少了對序列比對的依賴,增加了對殘基相互作用的重視。
- 限制:有時無法正確模擬分子的手性,無法預測大型蛋白質-核酸複合物的結構,生成模型可能會出現「幻覺」。
RNA 預測:
- AlphaFold 3 對 RNA 標靶的預測準確度高於其他方法,但不如頂級人類專家。
AlphaFold 伺服器:
- 提供使用者友善的介面,但原始程式碼和執行檔不公開。
- 偽代碼取代了原始碼,導致了爭論和阻礙了進一步的發展。
1. 在考慮AlphaFold 3 帶來的結構預測突破時,重要的是要記住,結構生物學的目標不是預測生物分子及其複合物的3D 結構,而是預測它們的行為以及執行生物程序時會發生什麼。
- 為了在預測分子行為方面取得進展,我們必須認識到結構預測問題並不像看起來那麼明確。生物分子及其複合物不會折疊成單一結構,而是形成數千種不同構象的集合,每種構像都有不同的機率和壽命。
- 了解這些構象景觀以及它們在生物分子相互作用時如何變化,對於定量預測親和力和動力學速率至關重要。
- 從各種條件下的序列預測構象集合是我們現在必須集中精力解決的問題,從而獲得對分子行為的定量和預測性理解。
- 儘管利用 AlphaFold 3 根據生物分子序列預測其自由和相互複合的 3D 結構,是理解分子行為和生物計算的重要一步,但實驗人員不必擔心被淘汰。結構生物學領域即將變得更加充滿活力。
論文連結:https://www.nature.com/articles/s41594-024-01350-2
The above is the detailed content of AlphaFold 3 takes an important step towards decoding molecular behavior and biological computing, a Nature sub-journal commented. For more information, please follow other related articles on the PHP Chinese website!

Hot AI Tools

Undresser.AI Undress
AI-powered app for creating realistic nude photos

AI Clothes Remover
Online AI tool for removing clothes from photos.

Undress AI Tool
Undress images for free

Clothoff.io
AI clothes remover

Video Face Swap
Swap faces in any video effortlessly with our completely free AI face swap tool!

Hot Article

Hot Tools

Notepad++7.3.1
Easy-to-use and free code editor

SublimeText3 Chinese version
Chinese version, very easy to use

Zend Studio 13.0.1
Powerful PHP integrated development environment

Dreamweaver CS6
Visual web development tools

SublimeText3 Mac version
God-level code editing software (SublimeText3)

Hot Topics
 1668
1668
 14
14
 1426
1426
 52
52
 1329
1329
 25
25
 1273
1273
 29
29
 1256
1256
 24
24
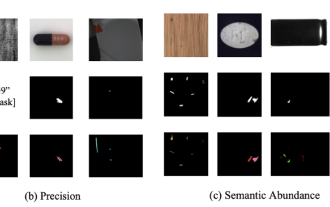 Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
Breaking through the boundaries of traditional defect detection, 'Defect Spectrum' achieves ultra-high-precision and rich semantic industrial defect detection for the first time.
Jul 26, 2024 pm 05:38 PM
In modern manufacturing, accurate defect detection is not only the key to ensuring product quality, but also the core of improving production efficiency. However, existing defect detection datasets often lack the accuracy and semantic richness required for practical applications, resulting in models unable to identify specific defect categories or locations. In order to solve this problem, a top research team composed of Hong Kong University of Science and Technology Guangzhou and Simou Technology innovatively developed the "DefectSpectrum" data set, which provides detailed and semantically rich large-scale annotation of industrial defects. As shown in Table 1, compared with other industrial data sets, the "DefectSpectrum" data set provides the most defect annotations (5438 defect samples) and the most detailed defect classification (125 defect categories
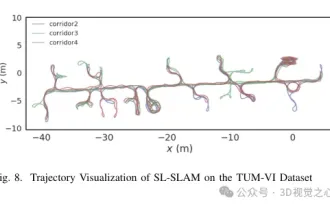 Beyond ORB-SLAM3! SL-SLAM: Low light, severe jitter and weak texture scenes are all handled
May 30, 2024 am 09:35 AM
Beyond ORB-SLAM3! SL-SLAM: Low light, severe jitter and weak texture scenes are all handled
May 30, 2024 am 09:35 AM
Written previously, today we discuss how deep learning technology can improve the performance of vision-based SLAM (simultaneous localization and mapping) in complex environments. By combining deep feature extraction and depth matching methods, here we introduce a versatile hybrid visual SLAM system designed to improve adaptation in challenging scenarios such as low-light conditions, dynamic lighting, weakly textured areas, and severe jitter. sex. Our system supports multiple modes, including extended monocular, stereo, monocular-inertial, and stereo-inertial configurations. In addition, it also analyzes how to combine visual SLAM with deep learning methods to inspire other research. Through extensive experiments on public datasets and self-sampled data, we demonstrate the superiority of SL-SLAM in terms of positioning accuracy and tracking robustness.
 Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Training with millions of crystal data to solve the crystallographic phase problem, the deep learning method PhAI is published in Science
Aug 08, 2024 pm 09:22 PM
Editor |KX To this day, the structural detail and precision determined by crystallography, from simple metals to large membrane proteins, are unmatched by any other method. However, the biggest challenge, the so-called phase problem, remains retrieving phase information from experimentally determined amplitudes. Researchers at the University of Copenhagen in Denmark have developed a deep learning method called PhAI to solve crystal phase problems. A deep learning neural network trained using millions of artificial crystal structures and their corresponding synthetic diffraction data can generate accurate electron density maps. The study shows that this deep learning-based ab initio structural solution method can solve the phase problem at a resolution of only 2 Angstroms, which is equivalent to only 10% to 20% of the data available at atomic resolution, while traditional ab initio Calculation
 NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
NVIDIA dialogue model ChatQA has evolved to version 2.0, with the context length mentioned at 128K
Jul 26, 2024 am 08:40 AM
The open LLM community is an era when a hundred flowers bloom and compete. You can see Llama-3-70B-Instruct, QWen2-72B-Instruct, Nemotron-4-340B-Instruct, Mixtral-8x22BInstruct-v0.1 and many other excellent performers. Model. However, compared with proprietary large models represented by GPT-4-Turbo, open models still have significant gaps in many fields. In addition to general models, some open models that specialize in key areas have been developed, such as DeepSeek-Coder-V2 for programming and mathematics, and InternVL for visual-language tasks.
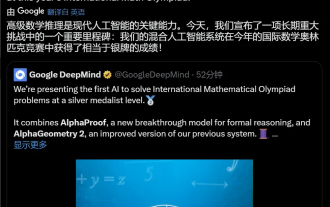 Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
Google AI won the IMO Mathematical Olympiad silver medal, the mathematical reasoning model AlphaProof was launched, and reinforcement learning is so back
Jul 26, 2024 pm 02:40 PM
For AI, Mathematical Olympiad is no longer a problem. On Thursday, Google DeepMind's artificial intelligence completed a feat: using AI to solve the real question of this year's International Mathematical Olympiad IMO, and it was just one step away from winning the gold medal. The IMO competition that just ended last week had six questions involving algebra, combinatorics, geometry and number theory. The hybrid AI system proposed by Google got four questions right and scored 28 points, reaching the silver medal level. Earlier this month, UCLA tenured professor Terence Tao had just promoted the AI Mathematical Olympiad (AIMO Progress Award) with a million-dollar prize. Unexpectedly, the level of AI problem solving had improved to this level before July. Do the questions simultaneously on IMO. The most difficult thing to do correctly is IMO, which has the longest history, the largest scale, and the most negative
 PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
PRO | Why are large models based on MoE more worthy of attention?
Aug 07, 2024 pm 07:08 PM
In 2023, almost every field of AI is evolving at an unprecedented speed. At the same time, AI is constantly pushing the technological boundaries of key tracks such as embodied intelligence and autonomous driving. Under the multi-modal trend, will the situation of Transformer as the mainstream architecture of AI large models be shaken? Why has exploring large models based on MoE (Mixed of Experts) architecture become a new trend in the industry? Can Large Vision Models (LVM) become a new breakthrough in general vision? ...From the 2023 PRO member newsletter of this site released in the past six months, we have selected 10 special interpretations that provide in-depth analysis of technological trends and industrial changes in the above fields to help you achieve your goals in the new year. be prepared. This interpretation comes from Week50 2023
 AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
AlphaFold 3 is launched, comprehensively predicting the interactions and structures of proteins and all living molecules, with far greater accuracy than ever before
Jul 16, 2024 am 12:08 AM
Editor | Radish Skin Since the release of the powerful AlphaFold2 in 2021, scientists have been using protein structure prediction models to map various protein structures within cells, discover drugs, and draw a "cosmic map" of every known protein interaction. . Just now, Google DeepMind released the AlphaFold3 model, which can perform joint structure predictions for complexes including proteins, nucleic acids, small molecules, ions and modified residues. The accuracy of AlphaFold3 has been significantly improved compared to many dedicated tools in the past (protein-ligand interaction, protein-nucleic acid interaction, antibody-antigen prediction). This shows that within a single unified deep learning framework, it is possible to achieve
 To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
To provide a new scientific and complex question answering benchmark and evaluation system for large models, UNSW, Argonne, University of Chicago and other institutions jointly launched the SciQAG framework
Jul 25, 2024 am 06:42 AM
Editor |ScienceAI Question Answering (QA) data set plays a vital role in promoting natural language processing (NLP) research. High-quality QA data sets can not only be used to fine-tune models, but also effectively evaluate the capabilities of large language models (LLM), especially the ability to understand and reason about scientific knowledge. Although there are currently many scientific QA data sets covering medicine, chemistry, biology and other fields, these data sets still have some shortcomings. First, the data form is relatively simple, most of which are multiple-choice questions. They are easy to evaluate, but limit the model's answer selection range and cannot fully test the model's ability to answer scientific questions. In contrast, open-ended Q&A





